We have performed genetic screens to discover novel mutants impacting on Drosophila metal homeostasis. One screen returned a mutant interacting with ferritin to reduce male fertility (Mehta et al., Biochemical Society Transactions, 2008), contributing to an understanding of the role of iron proteins during spermatogenesis (see also Metzendorf and Lind, BMC Developmental Biology, 2010, who showed that mitoferrin mutants cause male sterility). We have suggested a specific role of mitochondrial ferritin in testis function (Missirlis et al., Proceedings of the National Academy of Sciences U.S.A., 2006). Tharse Pathmanathan is characterizing this novel mutant and asks which are the critical requirements for iron during spermatogenesis.
➢ MOLECULAR PHYSIOLOGY OF METALS
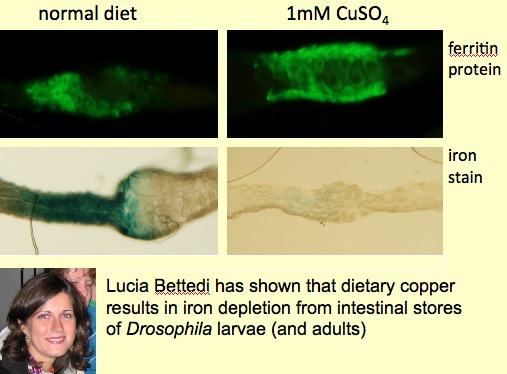
We propose that in the absence of a true liver, insects allocate iron storage to specific intestinal cells (Mehta et al., Biochimie, 2009). Regulation of ferritin in these cells is atypical. We are delineating iron trafficking pathways in Drosophila and discovered that dietary copper can mobilize iron from stores (Bettedi et al., The Journal of Experimental Biology, 2011), a finding observed and only partially understood in vertebrates, where a role for copper in the generation of haemoglobin is well documented.
We also reported a surprising difference in total zinc content between controls and fly models of Pantothenate Kinase Associated Neurodegeneration, a human disease known to affect iron accumulation in the brain of patients (Gutierrez et al., FEBS Letters, 2010). However, subsequent work showed the presence of an X-linked recessive mutation in most of our lab stocks (Afshar et al., FEBS Open Biology, 2013) and I am presently describing a molecular pathway that controls zinc storage in Drosophila melanogaster.
➢ EVOLUTION OF METAL HOMEOSTASIS
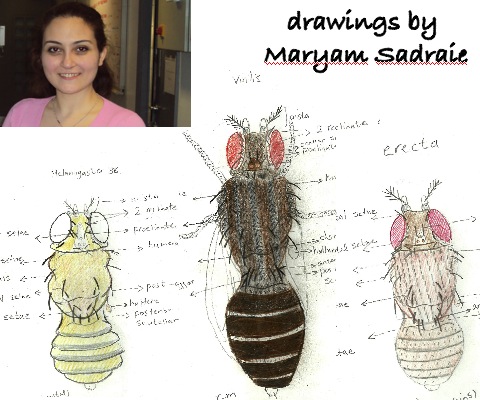
We are also working on evolutionary aspects of metal homeostasis (Sadraie and Missirlis, Biometals, 2011). We aim to exploit comparative genomics to explore and explain adaptations in metal regulation. For example, we have shown that although as a rule most species of Drosophila accumulate essential biological metals to similar extent, we could identify one notable exception for iron, which was found to accumulate in Drosophila erecta and to be scarce in Drosophila virilis. We could further show changes in ferritin accumulation in the intestines of these species. However, how ferritin is regulated remains unknown and we would like to use information from sequence comparisons between species to identify cis-regulatory elements that we could then characterize in Drosophila melanogaster. We are exploring experimentally the function of the conserved iron responsive element (Lind et al., JBC, 2006) present in the alternative exon of Fer1HCH.
Over the years, I have accumulated significant experience in oxidative stress and special strengths in the understanding of iron homeostasis and metabolism. I am developing further strategies to build on our understanding of metal biology as part of a systems research strategy to study the biological roles of metals. In particular, we are using Drosophila genetics to characterize novel cell biological pathways involved in metal physiology and homeostasis and are working to understand their function, integration and inter-dependence. Insights from the Drosophila model can inform both the understanding of human aging and the treatment of human disease.
To this end future research plans stem from ongoing projects in my lab:
➢ CELL BIOLOGY OF FERRITIN BIOSYNTHESIS
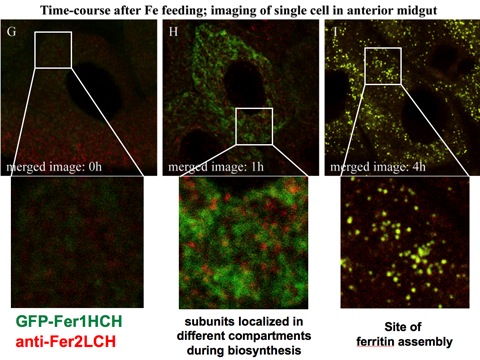
We have previously presented preliminary data showing that we could detect the individual ferritin subunits in discrete subcellular compartments prior to formation of the ferritin complex (see Figure adapted from Missirlis et al., Genetics, 2007). We now have both fly ferritin subunits tagged with fluorescent proteins and can follow the process in vivo. Our initial work suggests the ferritin assembly works differently depending on cell type. These findings complement and help explain the complexity we observed when we first studied ferritin in the adult brain (Kosmidis et al., Neurobiology of Disease, 2011). The work includes the use of novel photoconvertible forms of fluorescent proteins and confocal microscopy, in addition to the PACMAN technology developed by the Bellen lab (Venken et al., Nature Methods, 2009) for facilitating transgenesis of large genomic fragments. In our case of the entire locus encompassing both ferritin subunit genes, which are linked in Drosophila, was cloned and is being modified at will and then re-inserted in the fly.
➢ BIOPHYSICAL CHARACTERIZATION OF DROSOPHILA FERRITIN
Lucia Gutiérrez initiated this project in the lab and generated magnetic measurements from whole fly samples of different genotypes (for example ferritin mutants and transgenic overexpressors of ferritin). The magnetic properties can reveal iron speciation and include a clear ferritin signature in them. Jon Nield and Kristina Zubow performed single-particle electron microscopy reconstructions of ferritin from the same genotypes and our preliminary findings were recently reported (Gutiérrez et al., Metallomics, 2013)
➢ THE ROLE OF FERRITIN DURING EMBRYONIC DEVELOPMENT
I initiated this project during a six-month post-doctoral visit at the Institute of Neurology of the National Autonomous University of Mexico in the laboratory of Juan Riesgo Escovar. Nicanor Gonzales Morelos has been a close collaborator.
We have shown that lack of either ferritin gene product results in embryonic lethality. Mutant phenotypes range from non-deposition of cuticle to germ band retraction, dorsal closure and head involution defects. Female flies deposit different levels of ferritin in their embryos depending on variation in iron levels in the medium. We have also characterized embryonic nervous system phenotypes of ferritins, which include ventral nerve cord disruptions, misguided axonal projections and brain malformations. Ferritin mutants die with ectopic apoptotic events that partially explain nervous system phenotypes. (Gonzáles Morelos et al., PeerJ PrePrints, 2014)
➢ INTERPLAY BETWEEN BIOLOGICAL CLOCKS AND IRON HOMEOSTASIS
Konstantinos Mandilaras used Drosophila to assess potential molecular interactions between iron and the circadian clock machinery (Mandilaras and Missirlis, Metallomics, 2012). In the mammalian clock, heme is required to stabilize a key transcription factor involved in the molecular feedback loops that ensure proper periodic behaviour in the absence of external cues. 40 genes related to iron metabolism were silenced specifically in clock cells and the ability of such flies to sustain circadian rhythms in the absence of light cues was assessed. We screened for well-described behavioral alterations associated with mutations in the core molecular clock of Drosophila and have identified some genes implicated in iron metabolism that may also be required for the function of the central pacemaker. A remaining challenge is to identify precisely in which neuronal networks these iron-metabolism genes are required for proper circadian behaviour and the mechanism by which they affect the molecular clock.
➢ DROSOPHILA Fe-S CLUSTER AND MOLYBDENUM COFACTOR BIOSYNTHESIS
Most enzymes involved in the biosynthesis of Fe-S clusters and Molybdenum cofactor are encoded by human disease genes. Together with Helge Uhrigshardt we characterized Hsc20; a small chaperone involved in Fe-S cluster biogenesis (Uhrigshardt et al., Journal of Biological Inorganic Chemistry, 2013).
Silke Leimkühler’s group discovered that the L-cysteine desulfurase NFS1 (which has well-established functions in Fe-S cluster biosynthesis and in tRNA modification) a novel role of NFS1 in the sulfur transfer reaction for the biosynthesis of sulfur-containing molybdopterin (MPT), predicted to take place in the cytosol (Marelja et al., JBC, 2008). Zvonimir Marelja has performed an RNAi screen to identify missing components of the Molybdenum cofactor biosynthesis pathway in flies and in collaboration with Marina Georgiou, they have worked with the four Aldehyde Oxidase homologs revealed by the genome sequence (Marelja et al. J Exp. Biol., 2014).